About SM-TF
SM-TF database
The SM-TF database collects available 3D structures of small molecule-transcription factor complexes from Protein Data Bank (PDB). Totally, SM-TF contains 934 entries, covering 176 TFs from a variety of species. For each TF, SM-TF provides multiple conformations of binding pockets on the protein and also the structures complexed with different small molecules.
Transcription Factors
Database Setup
The structures of TFs were extracted from PDB using the following key words: "transcription factor", "transcriptional regulator", "transcriptional activator", "transcriptional repressor", "gene regulator", "gene activator", or "gene repressor". Only X-ray or NMR structures are kept in SM-TF database. Totally, 3077 PDB entries (July 3rd, 2015) were downloaded. The downloaded PDB entries were processed as follows:
Step 1
The PDB entries were grouped using the UniProt id of each protein. Proteins with the "sequence-specific DNA binding" function, according to the "Gene Orthology - molecular function" information provided by the UniProt database, were kept.
Step 2
Each PDB entry was searched for the HET information. The entries with only water molecules or ions were removed.
Step 3
The remaining entries were manually examined. Entries other than TFs were discarded.
Step 4
The remaining PDB entries were further reviewed. Entries containing functional small molecules were kept, and the entries containing only buffer or detergent ligands were removed. If there were more than one PDB entries containing an identical small molecule binding to the same pocket of the same protein, the structure with a higher resolution was kept.
Step 5
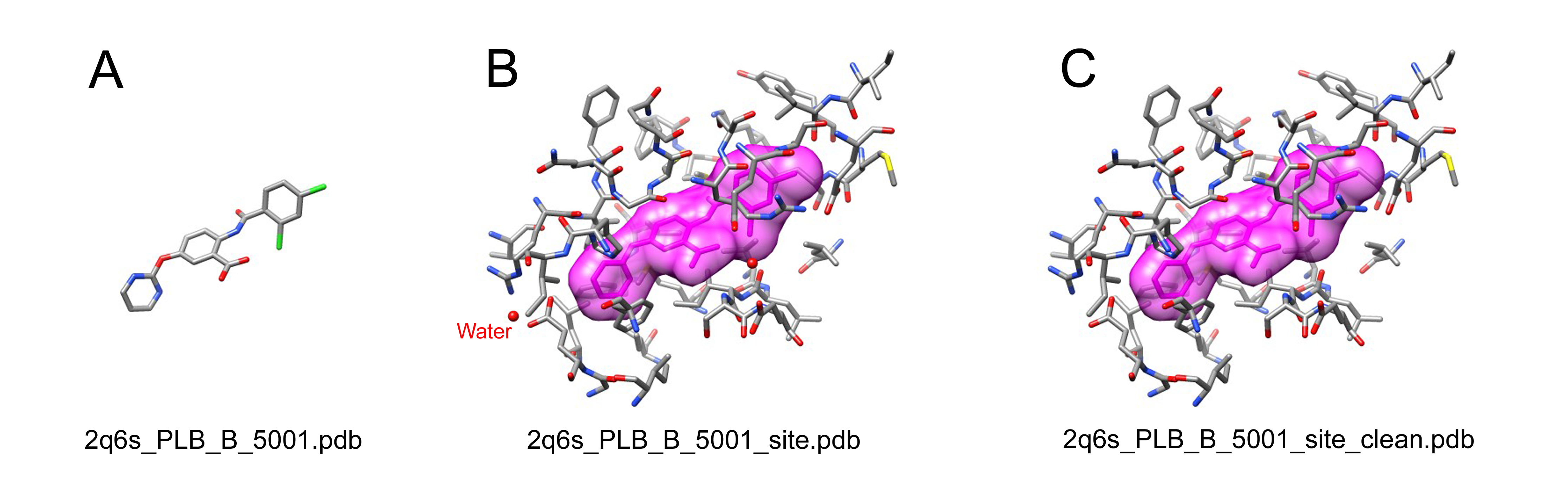
For each remaining entry, the small molecules of interest were extracted and named as "[PDB_id]_[HET_name]_[chain_id]_[resSeq].pdb". Amino acid residues and other ligands (including water molecules and ions, excluding the small moleculesaved in "[PDB_id]_[HET_name]_[chain_id]_[resSeq].pdb") within 6.5 Angstroms around the small molecule were defined as the binding site, named as "[small molecule file name]_site.pdb". Meanwhile, a pdb format file of the binding site containing only standard amino acid residues was created and named as "[small molecule file name]_site_clean.pdb".
Step 6
The TFs were categorized according to TF organisms and species.
Step 7
The data in the SM-TF database were linked to related databases such as UniProt, DrugBank, and other TF databases to provide detailed biological information.
Data Presentation
Example:
A: The structure of the small molecule, PLB;
Please see the following reference for more information:
Xu X, Ma Z, Sun H, Zou XQ.
SM-TF: A structural database of small molecule-transcription factor complexes.
Journal of Computational Chemistry , 37: 1559-1564, 2016.
[link]